Abstract
Background and Aims: Follicular lymphoma (FL) is an incurable B cell malignancy characterized by advanced stage disease and a heterogeneous clinical course. Despite recent genomic studies focused on profiling single FL biopsies over several time-points, little is known about the spatial heterogeneity as multi-site sampling is rarely conducted in lymphomas. However, in solid cancers, this approach has demonstrated profound spatial intra-tumor heterogeneity (ITH), impacting mechanisms of drug resistance and compromising precision-based strategies. As the majority of FL patients manifest disseminated tumor involvement, we sought to characterize the extent and clinical importance of spatial genetic heterogeneity in FL.
Methods: Our study cohort comprised 9 FL patients (SP1-9) from which 22 synchronously removed spatially-separated samples from several tissue sources (lymph node (LN), bone marrow (BM), skin (SK), pleural effusion (PE), ascites (AS), spleen (SP)) were collected. For patients SP3 and SP4, both spatial and longitudinal samples at 2 time-points; at FL and histological transformation, were included. FL B-cells and normal T-cells were enriched by fluorescence activated cell sorting (FACS) in most cases. Each sample was subjected to whole exome sequencing and we defined somatic variants, copy number alterations (CNAs) and mutation signatures using established bioinformatic tools. We employed a statistical clustering model (mclust) for clonal density estimation and together with ultra-deep targeted sequencing, reconstructed each tumor's subclonal architecture.
Results: Whilst spatially-separated FL tumors shared identical BCL2 - IGH breakpoints, we observed variable degrees of ITH. On average, 82% (range 50%-99%) of variants were shared between sites with two patterns of spatial ITH observed: high (SP1, SP8, SP9) and low (SP5, SP6, SP7). Copy number analyses showed a high level of concordance in most cases, except for SP4, SP7 and SP8. The overall distribution of known mutational signatures between spatially shared and discordant variants was different. Shared variants were predominantly enriched for mutational signature 1, representing age-related changes. In contrast signatures 6 and 24, both suggestive of defective DNA mismatch repair, were more pronounced in the spatially-discordant variants (χ2 test, p <0.0001), indicating that distinct mutagenic processes operate at different times during a tumor's evolution. Clonal architecture analyses revealed that some tumors comprised multiple competing subclones, with distinct clusters of mutations demonstrating differential expansions within spatially-separated sites, exemplified in SP2 (Figure 1). Importantly, for cases where spatial tumors were examined at two time-points, the degree of heterogeneity markedly increased with transformation. Detailed mutation analysis showed that apart from CREBBP mutations, that were clonally maintained throughout spatially separated biopsies, there were several instances of site-specific alterations in genes encoding proteins known to activate signaling pathways amenable to targeted therapies such as, TNFAIP3 (SP3 and SP9), TNFRSF14 (SP1), PIK3CD (SP4) and copy number loss of PTEN (SP8). In depth characterization of these variants using deep-sequencing techniques confirmed their discordant nature, suggesting ongoing genetic diversification driving evolution after widespread tumor dissemination.
Conclusion: Collectively, our results demonstrate that spatial ITH is prevalent in FL. The existence of site-specific aberrations suggests a single biopsy insufficiently captures the full genomic complexity present and these spatial variations need to be considered in biomarker-led clinical studies.
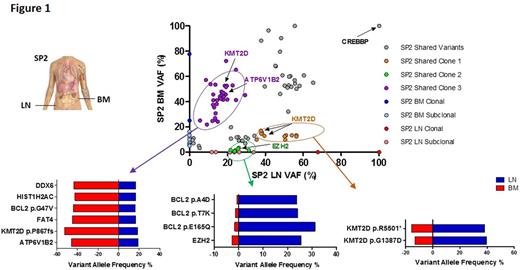
Figure 1: Dot plot demonstrating variants allele frequencies (VAFs) of all non-synonymous variants in SP2. Three distinct subclones circled in purple, orange and green can be inferred. The purple cluster includes ATP 6V1B2 and KMT2D variants classified as clonal in the BM and subclonal in the LN. The orange cluster includes different variants in KMT2D classified as subclonal in the BM and clonal in the LN. The green cluster includes variants in EZH2 and BCL2 that have a higher VAF in the LN compared to the BM. VAFs for selected variants in each of these putative subclones are shown in the horizontal bar graphs.
Auer: Janssen: Consultancy. Gribben: Kite: Honoraria; Karyopharm: Honoraria; Pharmacyclics: Honoraria; TG Therapeutics: Honoraria; Celgene: Honoraria; Acerta: Honoraria; Janssen: Honoraria; Genentech/Roche: Honoraria; Abbvie: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal